diurnal
diurnal is a Python library that facilitates RNA secondary structure
predictions.
This library aims at streamlining the process of elaborating, training, and validating RNA structure predictive models. Researchers can use it under the MIT license to develop and publish models that can be easily replicated by other users.
This page presents an Overview of the project and its Basic Usage. The User Guide provides detailed explanations of the library. The Source Code Documentation presents the signature of all the components of the library. The Literature Review provides the list of references that were used to develop the project and discusses other similar projects. The Developer Guide explains how the project is organized and how to contribute to it.
Overview
This library contains RNA secondary structure predictive models. It also comprises utility components that automate data processing tasks.
In short, RNA secondary structure describes the pairings of nucleotides. RNA (ribonucleic acid) is a molecule that performs a variety of biological functions. It is therefore the subject of interest for molecular biologists who aim at determining their function. An RNA molecules is made of a chain of nucleotides that can fold onto itself. One can describe the structure of RNA molecules in different ways.
The sequence of nucleotides is the primary structure. In general, RNA uses four possible nucleotides: adenine, cytosine, guanine, and uracil. They are represented by the letters
A,C,G, andU, respectively.The way that nucleotides combine with one another is the secondary structure. One way to represent the secondary structure is to use the dot-bracket notation. Unpaired nucleotides are represented by a dot (
.). A nucleotide paired with a nucleotide closer to the 3’ end of the molecule (i.e. to the right) is represented by an opening parenthesis ((). A nucleotide paired with a nucleotide closer to the 5’ end of the molecule (i.e. to the left) is represented by a closing parenthesis ()).The 3D arrangement of the molecule is the tertiary structure. The tertiary structure is not studied in this project but can be used to better understand the function of the molecule.
The image below displays the primary and secondary structures of a short RNA molecule.
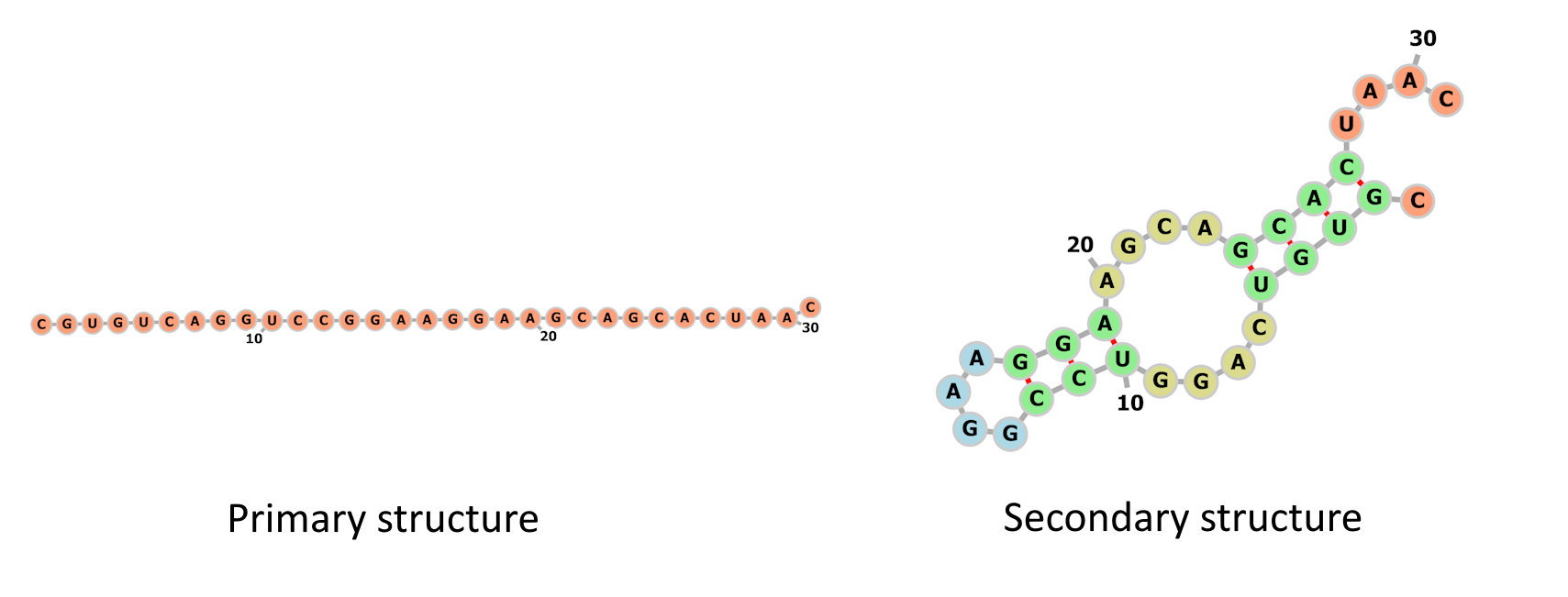
Primary and secondary structures of a short RNA molecule. Nucleotide sequence
taken from [1] and images generated with the forna
[2] visualization tool.
In the example above:
The primary structure can be represented as
CGUGUCAGGUCCGGAAGGAAGCAGCACUAAC.The secondary structure can be represented as
.((((....(((....)))....)))).....
Since determining the function of an RNA molecule from its primary structure is difficult, researchers rely on its secondary structure. Unfortunately, determining secondary structures experimentally is costly and time-consuming. There is hence an interest in reliably determining secondary structures from primary structures to understand the function of RNA molecules more effectively.
The diurnal library predicts secondary structures from primary
structures. It can:
download RNA structure datasets,
encode the datasets into trainable representations,
prepare the data for different evaluation methods,
train and evaluate models, and
visualize results.
diurnal is released under the MIT license and developed in Python. It relies
on the Numpy and PyTorch libraries for data manipulation and neural
network utilization.
Basic Usage
Consult the reference notebook at https://github.com/Vincent-Therrien/diurnal/blob/main/demo/example.ipynb for a complete use case of the library.